AI Revolutionizes Discovery of Mammalian Metabolites
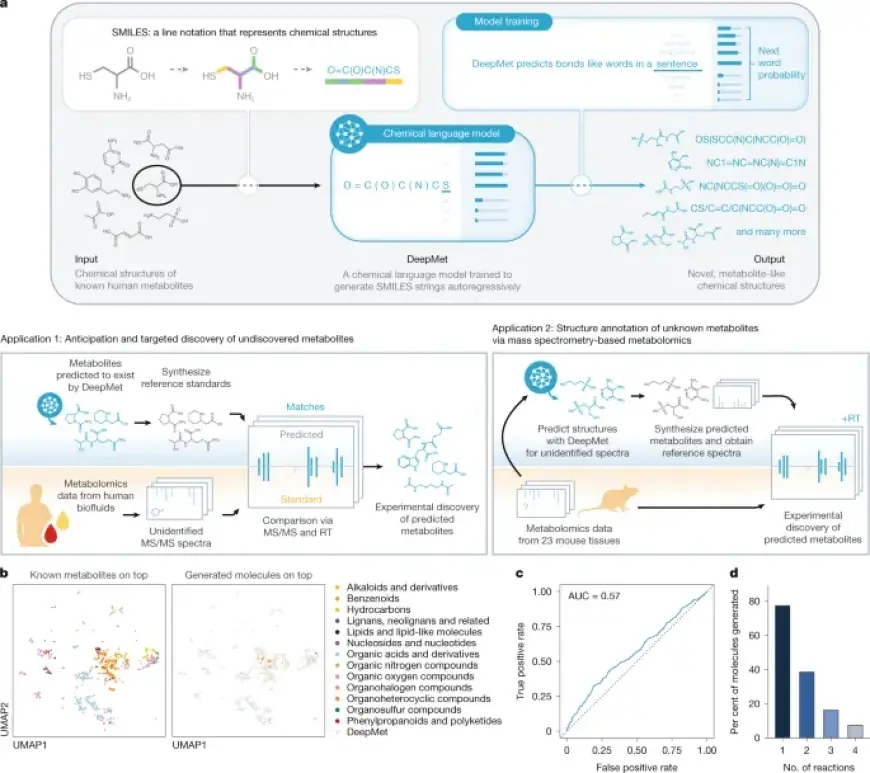
A recent study has highlighted a groundbreaking advancement in metabolomics, focusing on how artificial intelligence (AI) is transforming the discovery of mammalian metabolites. By utilizing machine learning techniques, researchers have developed a model named DeepMet, which enhances the identification and characterization of metabolites in mammalian systems.
AI-Driven Training Dataset Development
The foundation of this research relies on a training dataset sourced from the Human Metabolome Database (HMDB). This extensive database features 114,222 metabolites, with a majority classified as expected or predicted rather than experimentally verified. Out of these, only 8,970 metabolites were found to be detected or quantified, primarily consisting of lipids, which accounted for a significant number (6,791) of the identified compounds.
Data Filtering and Model Training
- Training set validation: To refine the dataset, lipids were excluded, resulting in a final training set of 2,046 small molecule metabolites.
- SMILES representation: The structures were converted into canonical SMILES format, ensuring data integrity for the language model.
- Model architecture: A recurrent neural network (RNN) based on long short-term memory (LSTM) was employed, demonstrating superior performance in generating metabolite-like structures.
Evaluating Metabolite Similarities
The researchers conducted various analyses to ensure that the AI-generated compounds exhibited characteristics akin to known metabolites. Techniques included dimensionality reduction to visualize chemical similarities and the use of a supervised machine learning classifier to differentiate between generated and actual metabolites.
Additionally, the authors utilized BioTransformer, a knowledgebase for enzymatic reactions, to further validate the generated metabolites against existing biotransformation products.
Integration with Experimental Data
To substantiate predictions made by DeepMet, a large-scale resource of anonymized human metabolomics data was leveraged. This resource facilitated validation through mass spectrometry, enhancing the reliability of the generated metabolites identified by the AI model.